Research
Organisms have had to evolve strategies to survive during fluctuating environmental conditions. Our research focuses on how cells are able to maintain homeostasis during these changing conditions by altering their gene expression. We have a number of projects that explore how gene expression is controlled, especially at the post-transcriptional level using the budding yeast Saccharomyces cerevisiae.
Current Projects
What is the mechanism and function of mRNA localization to the mitochondria?
Mitochondria are hubs for metabolite and energy generation and have been shown to be very important for age-related processes, including cancer and neurodegeneration, yet the mechanism of localized translation to these organelles and the impact this has on mitochondrial function is poorly understood. We found a novel mechanism of gene expression control where condition-dependent mRNA localization, regulated by translation elongation and the geometric constraints of the cell, controls protein synthesis of nuclear-encoded mitochondrial proteins. We are further expanding on our understanding of this means of gene expression control by generating quantitative measurements of translation elongation as well as building a computational model of mitochondrial mRNA localization that incorporate the changing mitochondrial morphology of the cell.
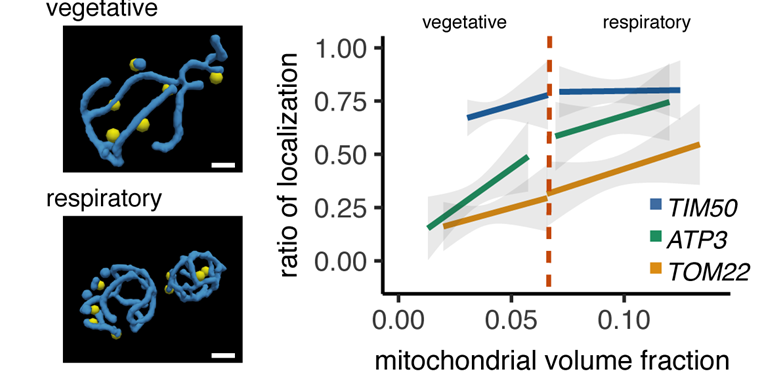
The fraction of the cytoplasm that is mitochondrial increases in respiratory conditions. ATP3 mRNA mitochondrial localization is especially sensitive to the mitochondrial volume fraction in its localization to the mitochondria.
How does the promoter specify the cytoplasmic fate of an mRNA during stress?
One potential mechanism for maintaining homeostasis during nutrient starvation is the transient formation of liquid-like phase transitions, which allows the cell to form membrane-less compartments that create intracellular partitions composed of dense concentrations of proteins and mRNAs. We have previously found that the promoter can direct the cytoplasmic fate of an mRNA at both the level of mRNA localization and translation. We are currently exploring the mechanism of this coupling between transcription and translation in eukaryotes.
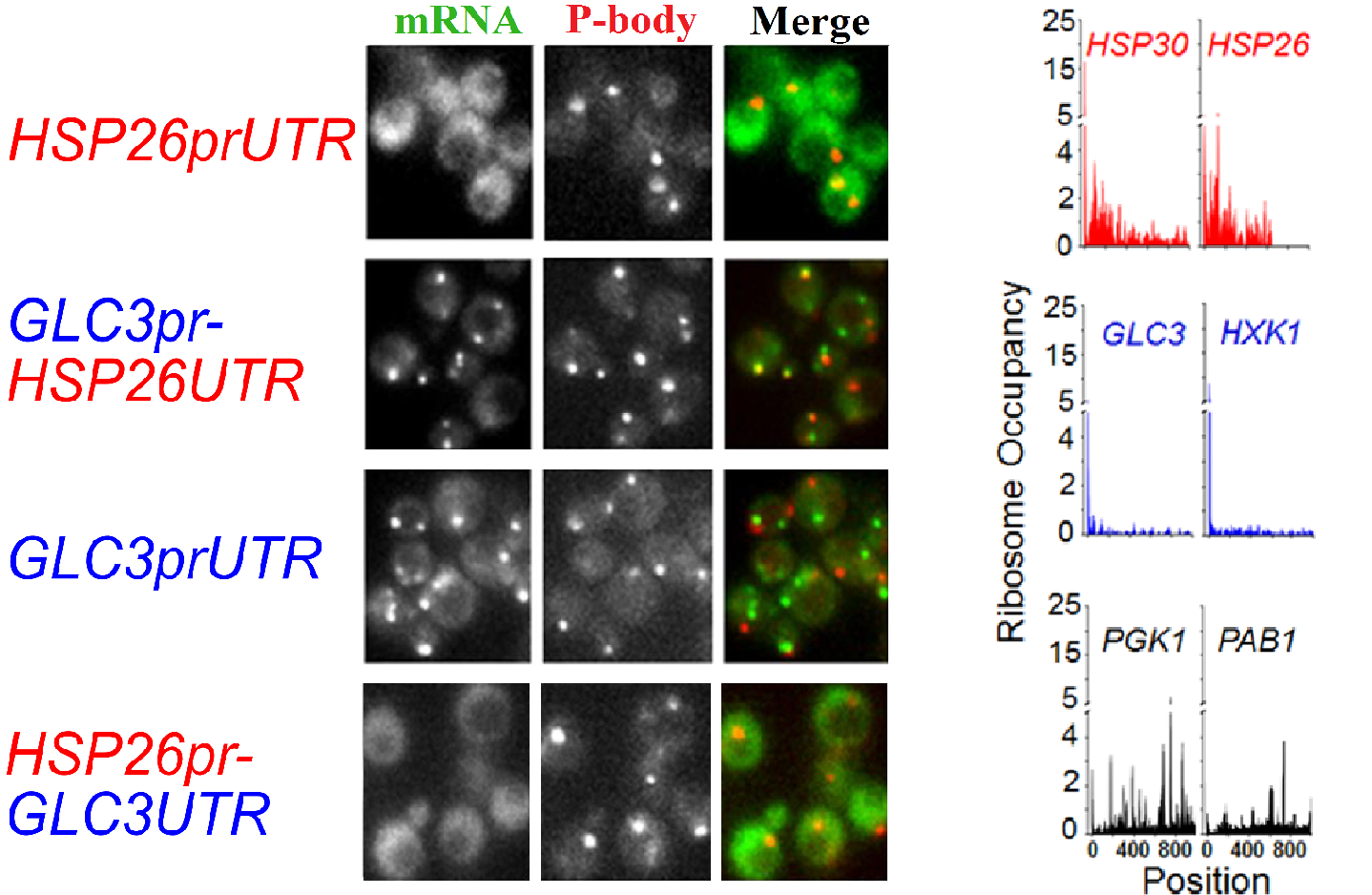
Transcriptionally upregulated, well translated HSP26 mRNAs remain diffusely localized during glucose starvation, while transcriptionally upregulated, poorly translated GLC3 mRNAs are sequestered to stress granules. In both cases the promoter is the determining factor in these localization and translation phenotypes. While ribosome profiling allows us to measure codon level resolution of the distribution of ribosomes accross mRNAs in glucose starvation.
What happens to aggregation prone RNA-binding proteins during aging?
Age-related neurodegenerative diseases are a class of incurable diseases that result in the progressive degeneration of neuronal cells. Recent research has pointed to a role for RNA-binding proteins (RBPs) in age-related neurodegenerative diseases, as many RBPs have aggregation-prone intrinsically disordered regions, they accumulate in pathological inclusions of various neurodegenerative diseases, and mutations in a number of RBPs have been linked to neurodegenerative diseases. We are currently exploring the aggregation of yeast RNA-binding proteins during aging in yeast.